
|
ConfeitoGUI |
a tool for correlation network analysis |
★ About ConfeitoGUI
♦ Abstract
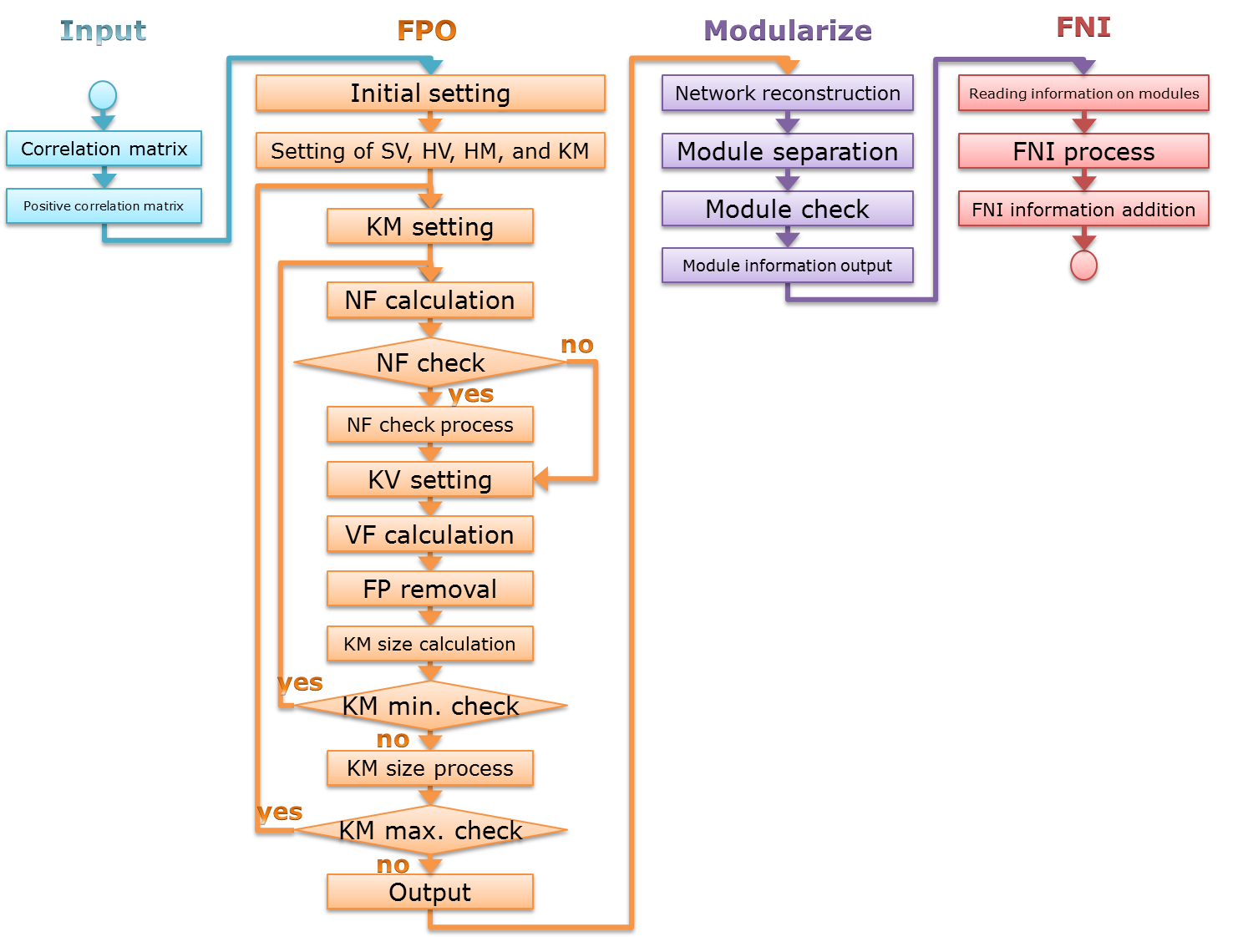
ConfeitoGUI is a stand-alone tool for extracting local communities (or network module in this site),
based on the Confeito algorithm.
The Confeito algorithm is an algorithm to extract network modules from a correlation network.
♦ Usage
-
Install of Java Runtime Environment (JRE)
-
Check the version of JRE: "java -version" on the command prompt in Windows or the terminal in Linux
(Caution: ConfeitoGUI is performable only on the 64-bit version of JAVA)
- 64-bit version of JRE is available at here.
- For Windows users, click the “Windows Offline (64-bit)” to install the package.
- For Linux users, refer to the site.
-
Install of ConfeitoGUI
- Expand the file "ConfeitoGUI_x.x.x.zip" into a folder.
- The tool is available here.
-
Executionof ConfeitoGUI
- Doublclick the file "launch.bat" expanded above to execute the tool.
- To know how to analyze using the tool, see the page of example 1.
♦ Steps for analysis
-
Array Data Converter (optional):
to change raw data of microarray experiments by Affymetrix and Agilent into a text-formatted file.
-
Correlation Tool:
to create a correlation matrix between elements. Cosine and Pearson correlation coefficients are selectable.
-
Preprocessing Tool:
to make a two-column list composed of identifiers of elements and names of files including correlation data by elements.
- Check Tool: to check files of correlation data.
-
FPO (False-Positive-Out) analysis:
to select out elements that are highly and excusively correlated to each other
and thus, are composed of a network module.
-
Among elements with high correlation coefficients to an element,
there can be some elements with high correlation to any (or a lot of) elements;
namely “false-positive” elements.
- ConfeitoGUI can efficiently remove out such false-positive elements from a network module.
-
FNI (False-Negative-In) analysis (optional):
for a network module, to select out elements that are specifically related to the module,
in spite of low correlation coefficient to the eleements included in the module,
namely “false-negative” elements.
-
Using tools for detecting local communities from a network including the FPO analysis,
it is difficult to detect such elements, because of low correlation coefficients to elements of the module.
-
By combining the FPO and FNI analyses,
ConfeitoGUI can remove out false-positive elements and select out false-negative elements.
|
 |
♦ Comparison with the other tools for community detection
To check the accuracy of network modules that are extracted from ConfeitoGUI,
we obtained a dataset of mouse microarray experiments from Gene Expression Omnibus (GEO), NCBI
and compared ConfeitoGUI with the other tools for community detection in their memberships of network modules.
-
About GEO
- GPL: A platform of gene expression datasets, e.g., microarray and next-generation sequencing.
- GSE: Experiments included in GPL.
- GSM: Gene expression data belonged to GSE.
- In many cases, experiments included in a GSE are similar to each other in their gene expression data.
-
Namely, experiments (GSMs) in a GSE are expected
to belong to the same network module obtained by tools for community detection.
-
A dataset, calculation of correlation coefficient, and depiction of correlation network
-
A dataset of 37,013 chips of Affymetrix mouse microarray
(GPL1261) was used (a list of the dataset is available at here).
- Cosine correlation coefficients were calculated between experiments to make a correlation matrix.
- We depicted correlation networks based on the matrix from 0.50 to 0.99 (0.01 step) as thresholds of the networks.
-
The other tool for community detection to be compared
- Louvain (Blondel et al., J Stat Mech, 2008):
Executable on the Pajek software.
- Simulating annealing (Newman and Girvan, Phys Rev E, 2004):
Executable on R.
- Fast greedy (Clauset et al., Phys Rev E, 2004):
Executable on R.
-
Methods: to check similarity of memberships between network module and GSE experiments
- F-measure was used for the check.
-
In the field of information retrieval,
F-measure, hamonic mean of precision and recall.
-
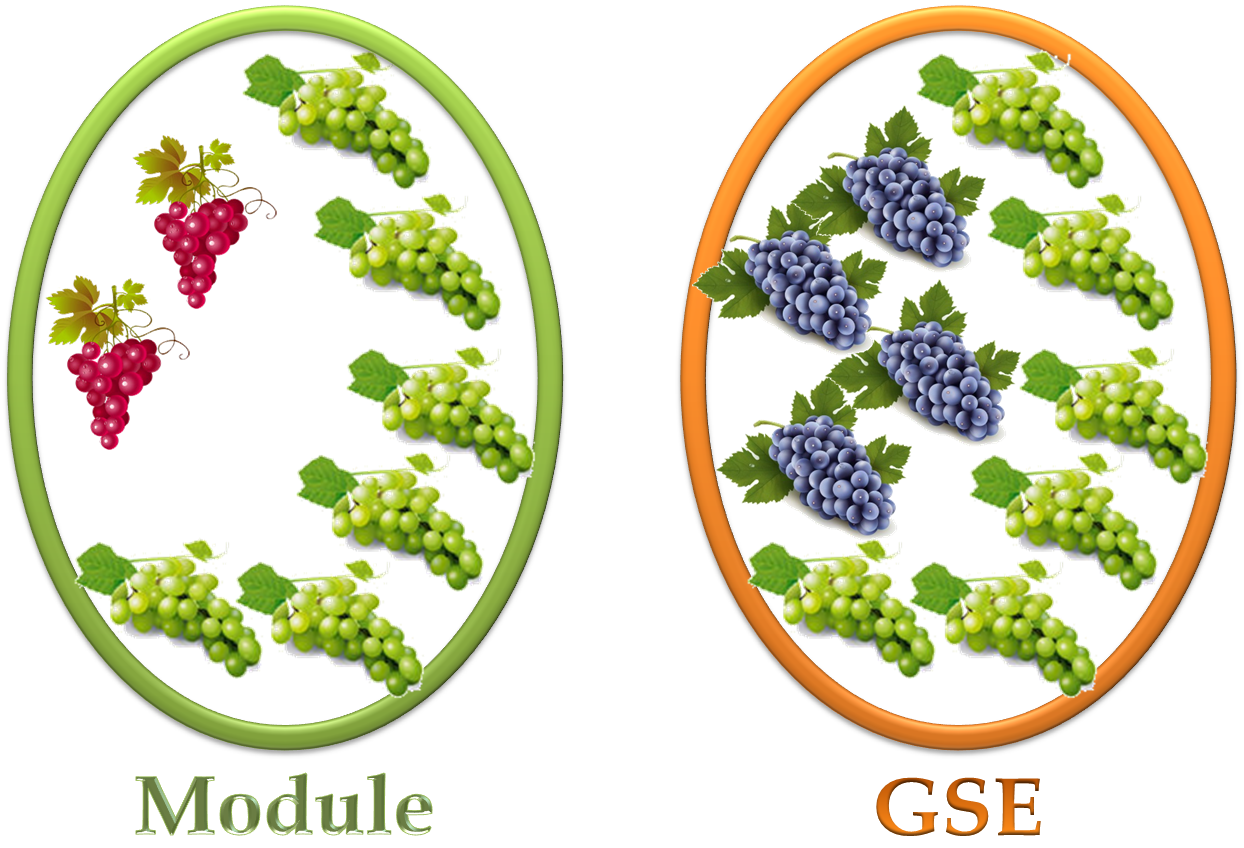
The precision index here is the proportion of experiments
shared with a network module and a GSE (
 )
over all experiments in the module ( )
over all experiments in the module (  ). ).
-
The recall index here is the proportion of the shared experiments (
 )
over all experiments in the GSE ( )
over all experiments in the GSE (  ). ).
-
In the lower figure, the precision index is 6 / 8 = 0.75,
and the recall index is 6 / 10 = 0.60.
-
Namely, The value of F-measure is ( 6 + 6 ) / ( 8 + 10 ) = 0.67.
- The larger F-meausre, the more similar in the memberships of a network module and a GSE.
- The values of F-measure were calculated for all network modules in the correlation networks with various thresholds.
- For each correlation network, an average value of F-measure was calculated to compare the values between the tools.
- Namely, the higher values of average F-measure, the more accurate network modules.
|
|
-
Results
- The lower table represents the average values of F-measure of the tools.
| Tool | F-measure (average ± SD) |
| ConfeitoGUI | 0.61 ± 0.34 |
| Louvain | 0.47 ± 0.40 |
| Simulating annealing | 0.40 ± 0.36 |
| Fast greedy | 0.37 ± 0.37 |
- The average value of F-measure of ConfeitoGUI is the highetst and thus, the tool shows the accuracy of network modules.
♦ References
-
Ogata Y et al., ConfeitoGUI: a toolkit for community detection from a correlation network in molecular biology,
with a function to adjust community size. (submitting)
-

 Sakurai N et al., KaPPA-View4: a metabolic pathway database for representation and analysis of correlation
networks of gene co-expression and metabolite co-accumulation and omics data.
Nucleic Acids Res, 39: D677-D684, 2011.
Sakurai N et al., KaPPA-View4: a metabolic pathway database for representation and analysis of correlation
networks of gene co-expression and metabolite co-accumulation and omics data.
Nucleic Acids Res, 39: D677-D684, 2011.
-

 Ogata Y et al., CoP: a database for characterizing co-expressed gene modules
with biological information in plants. Bioinformatics, 26(9): 1267-1268, 2010.
Ogata Y et al., CoP: a database for characterizing co-expressed gene modules
with biological information in plants. Bioinformatics, 26(9): 1267-1268, 2010.
-

 Ogata Y et al., The prediction of local modular structures in a co-expression network
based on gene expression datasets. Genome Inform, 23(1): 117-127, 2009.
Ogata Y et al., The prediction of local modular structures in a co-expression network
based on gene expression datasets. Genome Inform, 23(1): 117-127, 2009.
♦ The advanced version of ConfeitoGUI
The version in this site is an academic-free version. About the advanced (or full) version of ConfeitoGUI,
ask Kazusa DNA Research Institute.
Back to the portal.
 |
|
The KAGIANA Project |
|
(since 2006) |
Copyright (C) 2015 All rights reserved Osaka Prefecture University
& Kazusa DNA Research Institute

