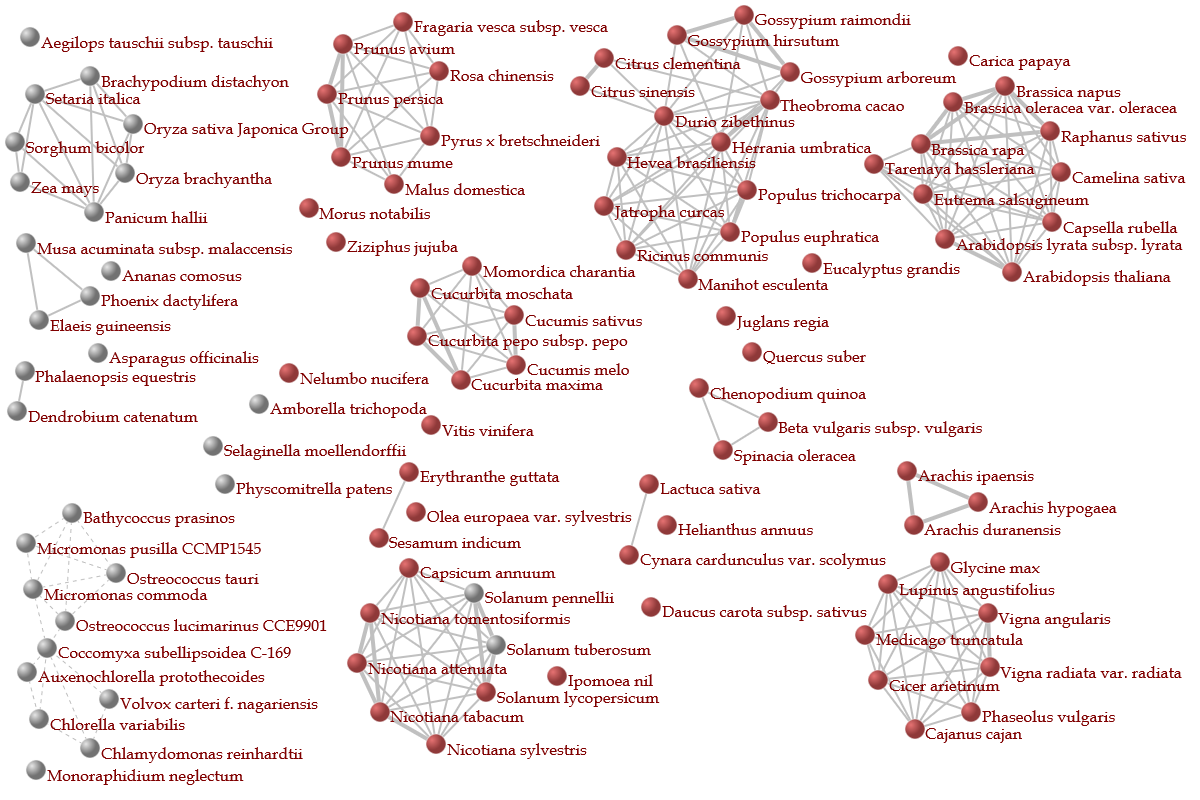
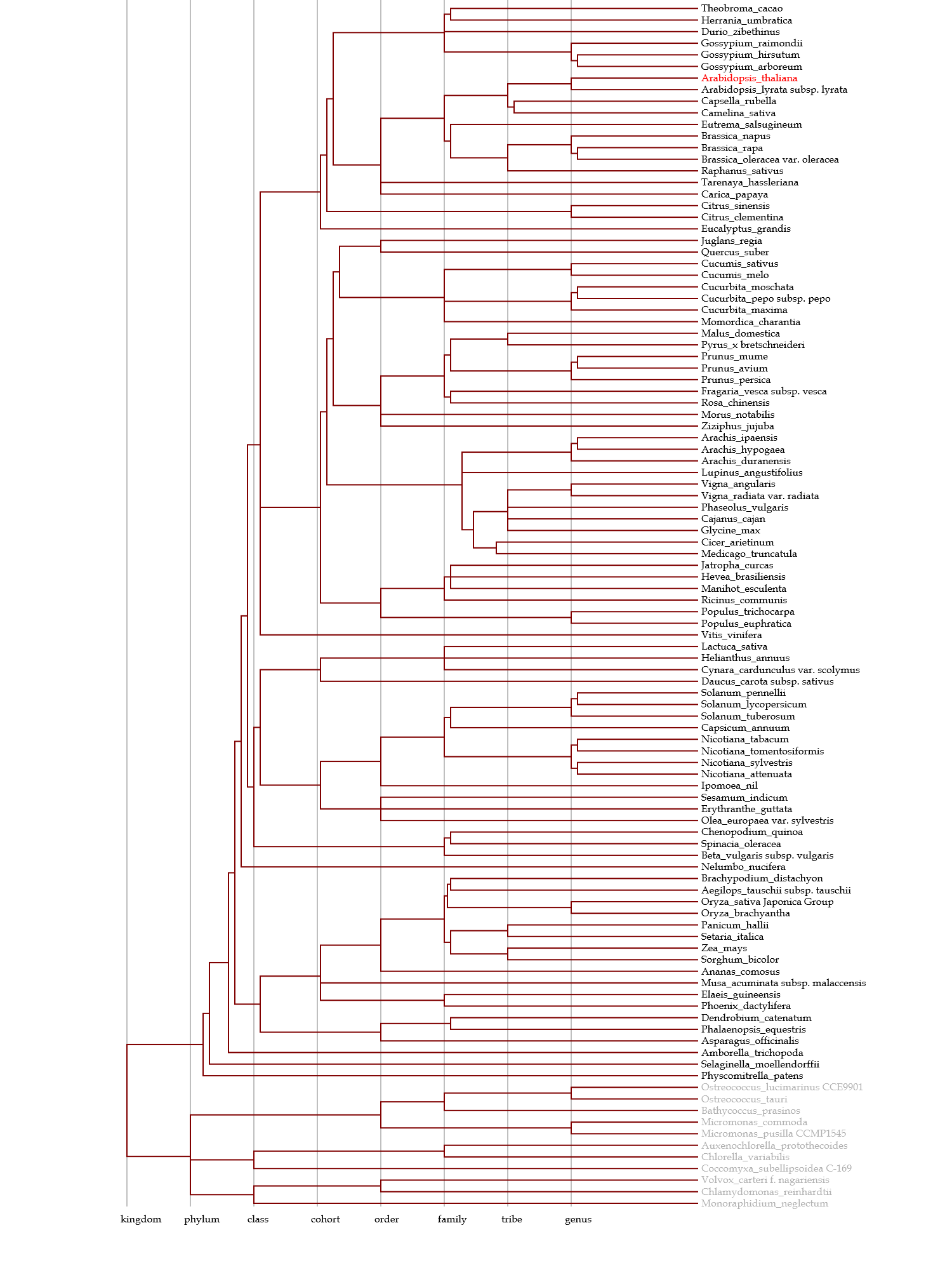
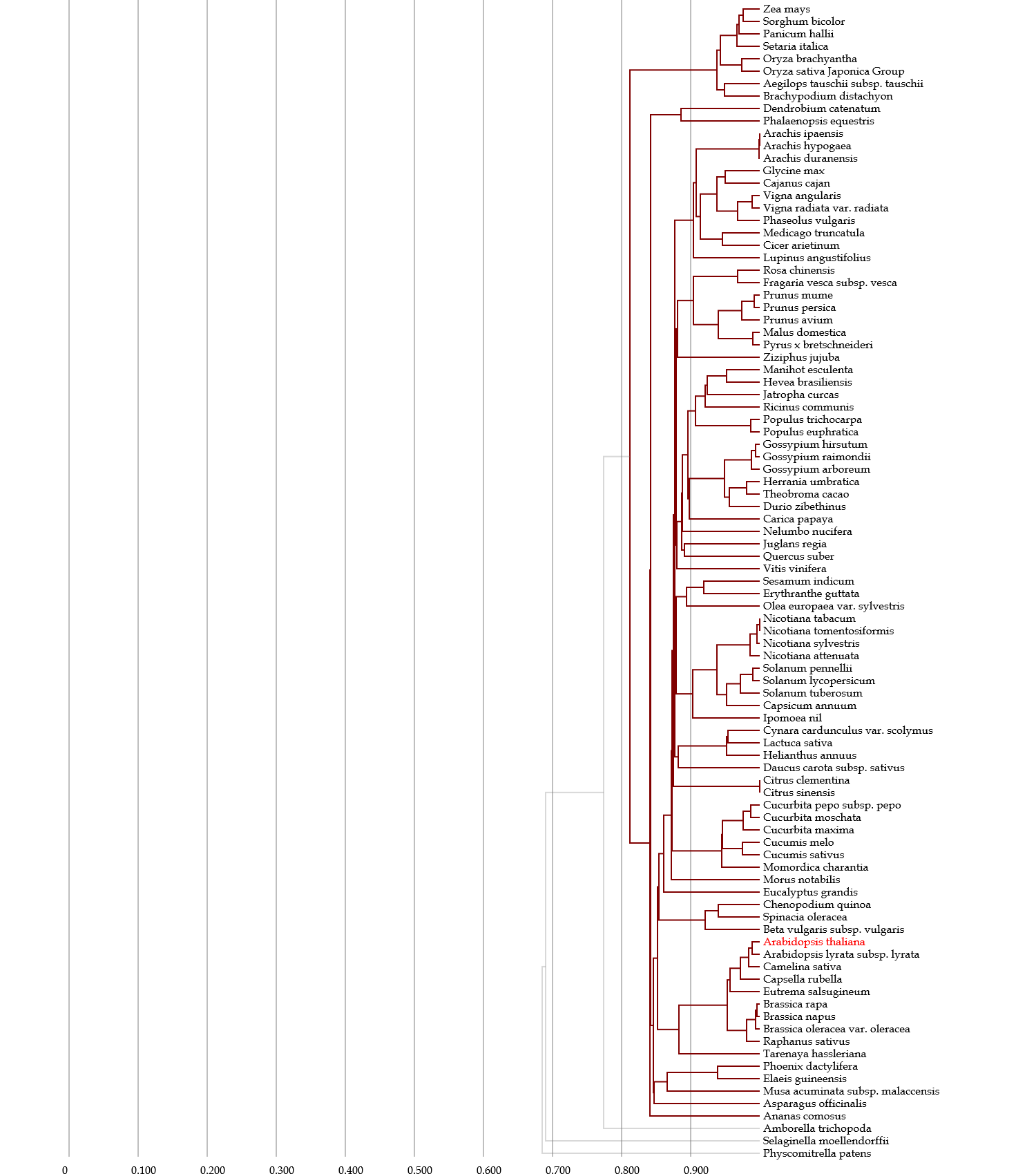
- This network contains nodes (organisms), and these nodes are interconnected to each other.
-
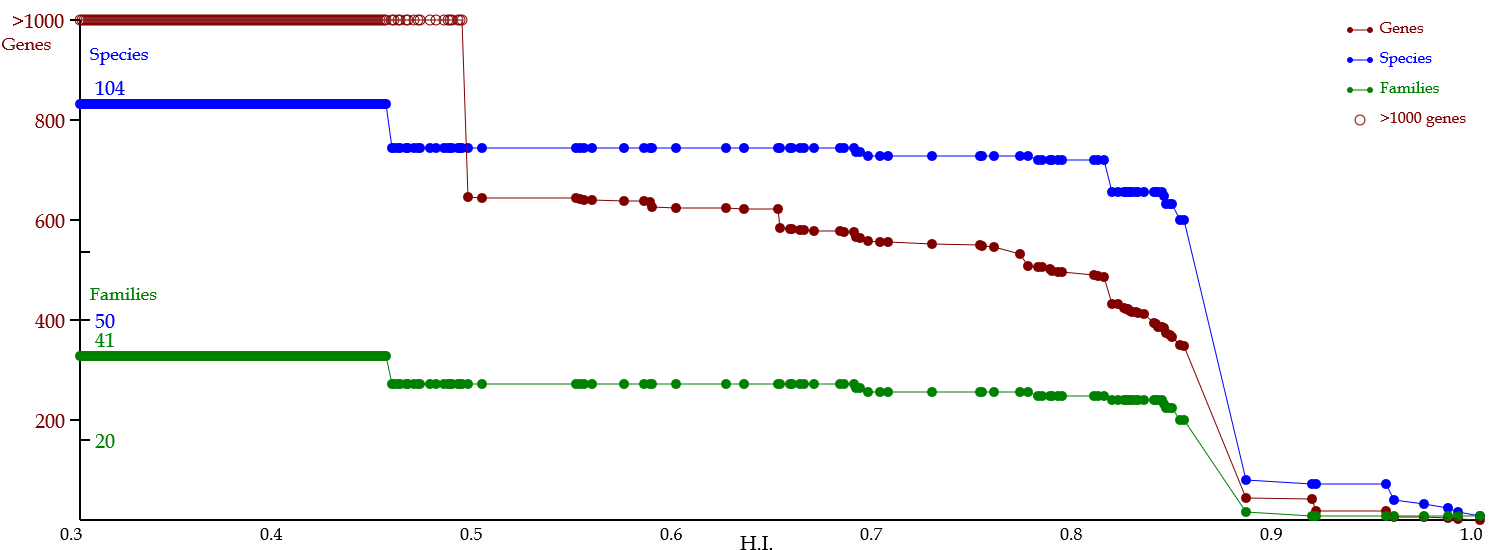
For the interconnection, the C.I. value was used as a threshold.
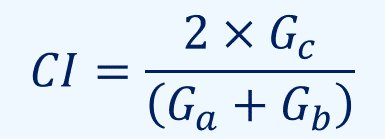
-
In the above equation, Ga, Gb, & Gc represent
the numbers of genes contained in Organism A, contained in Organism B, & shared between these organisms.
-
The index ranges from zero to one.
-
When the index is one, a pair of organisms perfectly share their homologous genes,
& when it is zero, the organisms share no homologous genes.
-
The topology of the organisms is based on the indices, but their positions are arbitrary.
-
In the network, a network module, in which nodes are tightly interconnected to each other,
hypothetically represents an organism group that share many homologous genes with each other,
and also less homologous genes with the other organisms.
-
In plants, 0.6 of the threshold was used, because several families are separated as network modules.
|
|